Prepare your data
Set your genome with useGenome()
useGenome('hg19')
#> Genome has been set to hg19
retrieveGenome()
#> Retrieving: hg19
#> # A tibble: 33,575 x 8
#> symbol start_position end_position chromosome_name arm band strand
#> <chr> <dbl> <dbl> <fct> <fct> <chr> <int>
#> 1 DDX11… 11869 14412 1 1p p36.… 1
#> 2 WASH7P 14363 29806 1 1p p36.… -1
#> 3 MIR13… 29554 31109 1 1p p36.… 1
#> 4 FAM13… 34554 36081 1 1p p36.… -1
#> # … with 33,571 more rows, and 1 more variable: ensembl_gene_id <chr>Infer CNAs and plot a heatmap
Infer cells’ CNA profiles with infercna()
args(infercna)
#> function (m, refCells = NULL, window = 100, range = c(-3, 3),
#> n = NULL, noise = 0.1, center.method = "median", isLog = FALSE,
#> verbose = TRUE)
#> NULL
cna = infercna(m = m, refCells = refCells, n = 5000, noise = 0.1, isLog = TRUE, verbose = FALSE)
cnaM = cna[, !colnames(cna) %in% unlist(refCells)]
Plot heatmap with cnaPlot()
args(cnaPlot)
#> function (cna, limits = c(-1, 1), ratio = 0.5, cols = heatCols,
#> x.name = "Chromosome", y.name = "Cell", legend.title = "Inferred CNA\n[log2 ratio]",
#> x.hide = c("13", "18", "21", "Y"), orderCells = F, order.with = NULL,
#> euclid.dist = F, angle = NULL, x.angle = NULL, y.angle = 0,
#> axis.rel = 1, base.size = 12, axis.title.size = 12, axis.text.size = 11,
#> base.col = "#073642", title = NULL, subtitle = NULL, caption = NULL,
#> text.size = 12, y.hide = NULL, tile.size = 0.1, tile.col = NULL,
#> legend.position = "right", legend.height = 2, legend.width = 0.6,
#> legend.rel = 0.9, legend.colour = "black", legend.breaks = NULL,
#> legend.labels = NULL, legend.justification = "top", legend.title.position = "bottom",
#> legend.title.angle = NULL, legend.title.rel = 0.9)
#> NULL
obj = cnaPlot(cna = cnaM,
orderCells = TRUE,
subtitle = 'Copy-Number Aberrations in a patient with Glioblastoma')
names(obj)
#> [1] "p" "data"
obj$data
#> # A tibble: 1,665,660 x 3
#> Gene Cell CNA
#> <fct> <fct> <dbl>
#> 1 WASH7P MGH125-P2-H01 0
#> 2 NOC2L MGH125-P2-H01 0
#> 3 ISG15 MGH125-P2-H01 0
#> 4 SDF4 MGH125-P2-H01 0
#> # … with 1,665,656 more rows
obj$p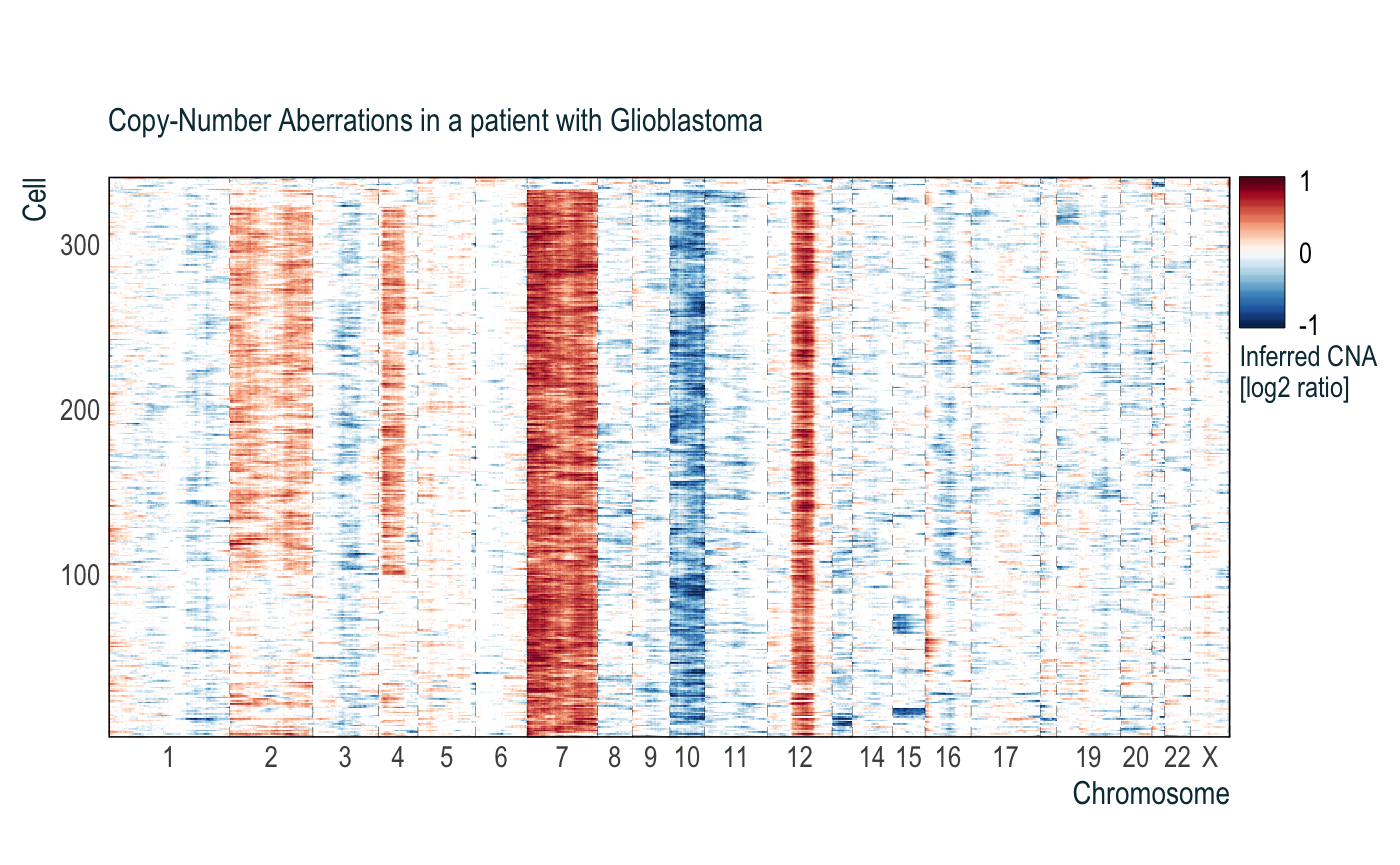
Ordering cells in cnaPlot()
In cases where you can see the subclones in the heatmap but don’t yet have the subclone assignments to direct the cell clustering, you can instead give a set of chromosome and chromosome arms that you would like the clustering to use. In the example above, we probably want the cells belonging to the presumed subclones (chromosome 2, chromosome arm 4p, etc) to be grouped together. We can specify this using the order.with argument in infercna::cnaPlot:
obj = cnaPlot(cna = cnaM,
orderCells = TRUE,
order.with = c(2, "4p", 7, 10, 12, 13, 15),
subtitle = 'Copy-Number Aberrations in a patient with Glioblastoma')
obj$p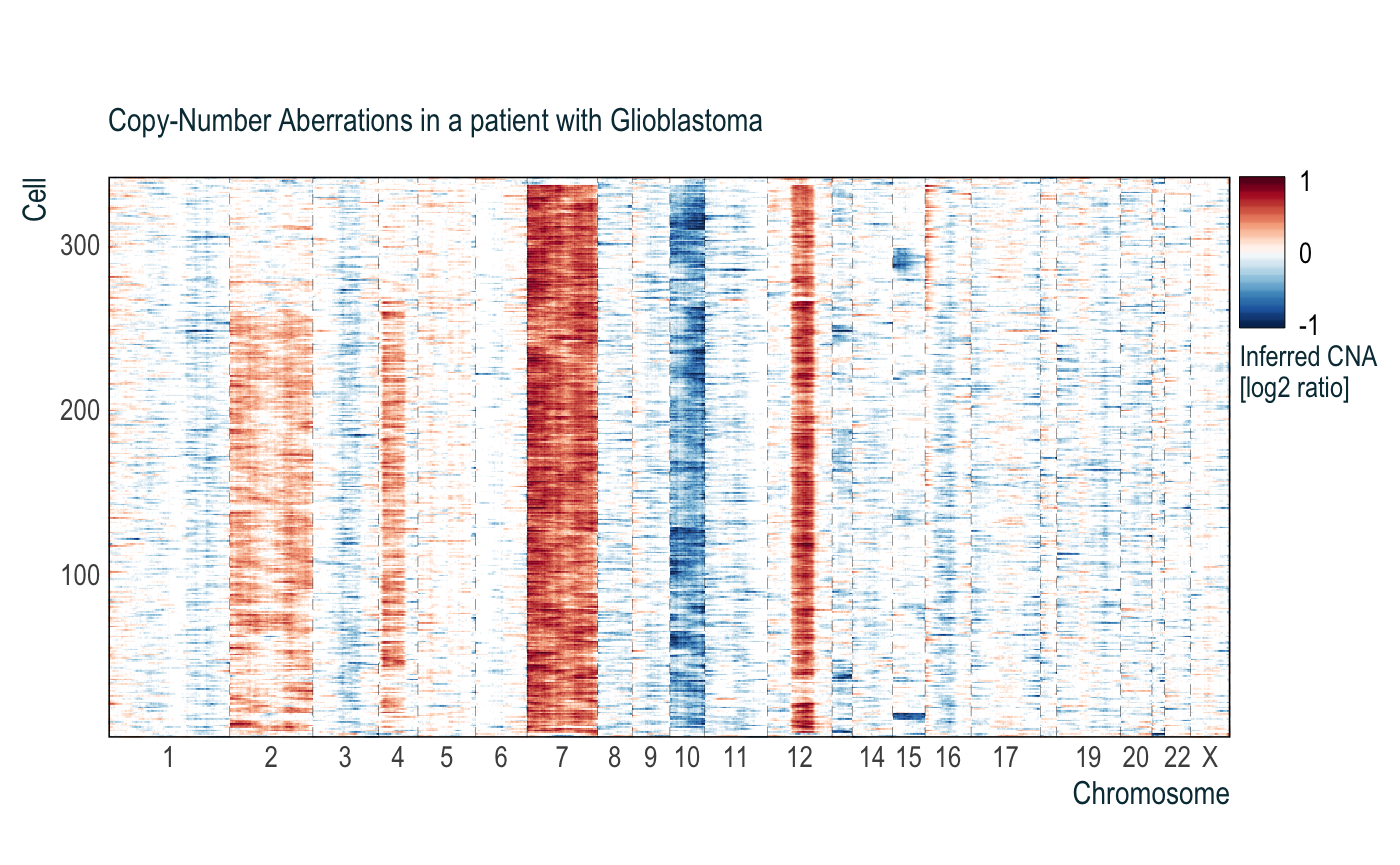
Find malignant cells
In the above section, we filtered out the infercna::refCells that we know are normal prior to looking at the CNA heatmap. In reality, our reference normal cells will only comprise a subset of all normal cells in the data, and we will need to identify the remaining ones to exclude them from downstream analyses concerning the malignant cells only.
infercna defines two parameters, infercna::cnaCor and infercna::cnaSignal that quantify the extent of copy-number aberrations in individual cells and thus help to separate the malignant and non-malignant subsets.
Compute cnaSignal() and cnaCor()
cnaSignal()
CNA signal reflects the overall extent of CNAs. It’s defined as the mean of the squares of CNA values across the genome and should therefore accentuate genome-wide differences in CNA profiles between malignant and non-malignant cells.
cnaCor()
CNA corrleation refers to the correlation between the CNA profile of each cell and the average CNA profile of all cells. Best results are seen when correlating cells to the average CNA profile of cells from the corresponding tumour and, if possible, excluding from the average those already classified as non-malignant.
Visualise CNA Signal and Correlation with cnaScatterPlot()
Plotting cnaCor against cnaSignal for all cells is a good first approximation to the malignant and non-malignant groups. infercna::cnaScatterPlot simply integrates these two function calls and their respective parameters into one function, and plots the results.
args(cnaScatterPlot)
#> function (cna, cor.method = "pearson", threshold = NULL, cor.threshold = threshold,
#> signal.threshold = threshold, group = NULL, group.col = "magenta",
#> cex = 0.5, hline = NULL, vline = NULL, bySample = FALSE,
#> samples = NULL, sep = "-|_", excludeFromAvg = NULL, ...)
#> NULLLet’s quickly go over the note-worthy arguments:
-
<threshold>: of which genes to include in calculation; it determines what fraction of genes with top CNA signal to keep.- in
cnaSignal() - in
cnaCor()
- in
-
<cor.threshold>: inherits from<threshold>; can be supplied if the threshold intended is specific toinfercna::cnaCorcalculation.- in
cnaCor()
- in
-
<signal.threshold>: inherits from<threshold>; can be supplied if the threshold intended is specific toinfercna::cnaSignalcalculation.- in
cnaSignal()
- in
-
<samples>: used to determine tumour-specific CNA profiles; can be one of:- names of tumour samples (unique)
- vector of tumour samples of length equal to the number of columns and ordered correspondingly.
-
<excludeFromAvg>: names of the (normal) cells to be excluded from the average CNA profile(s).
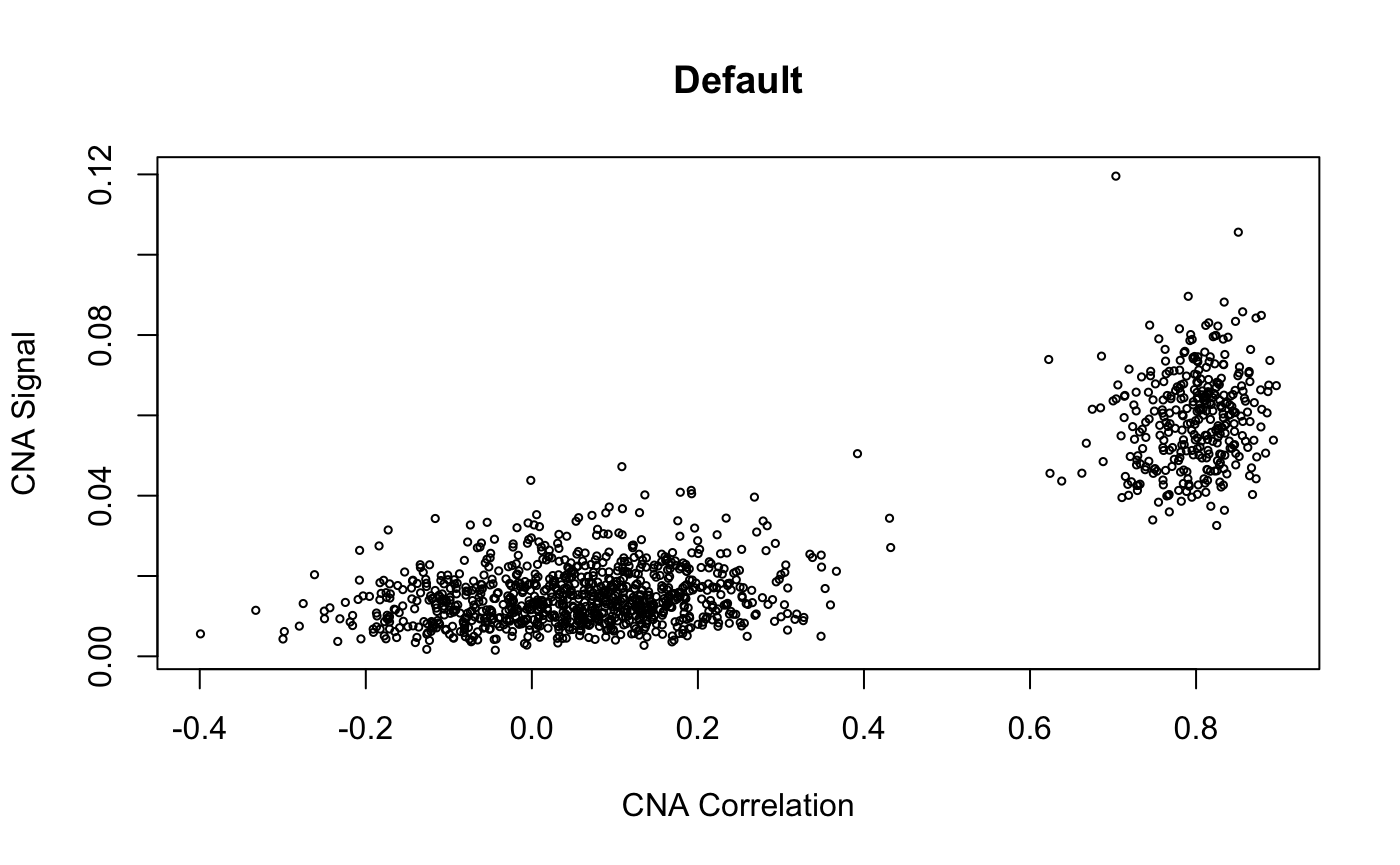
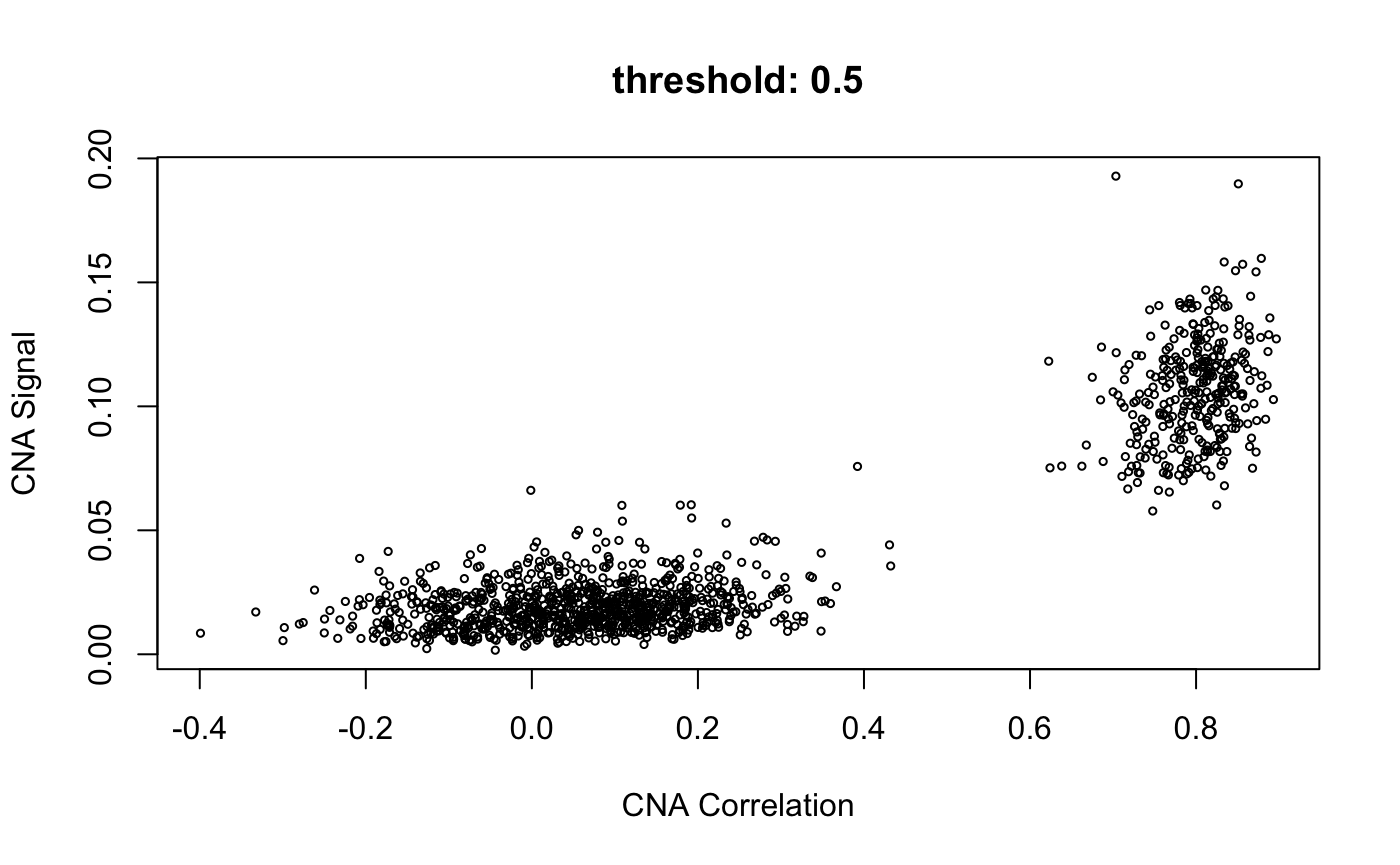
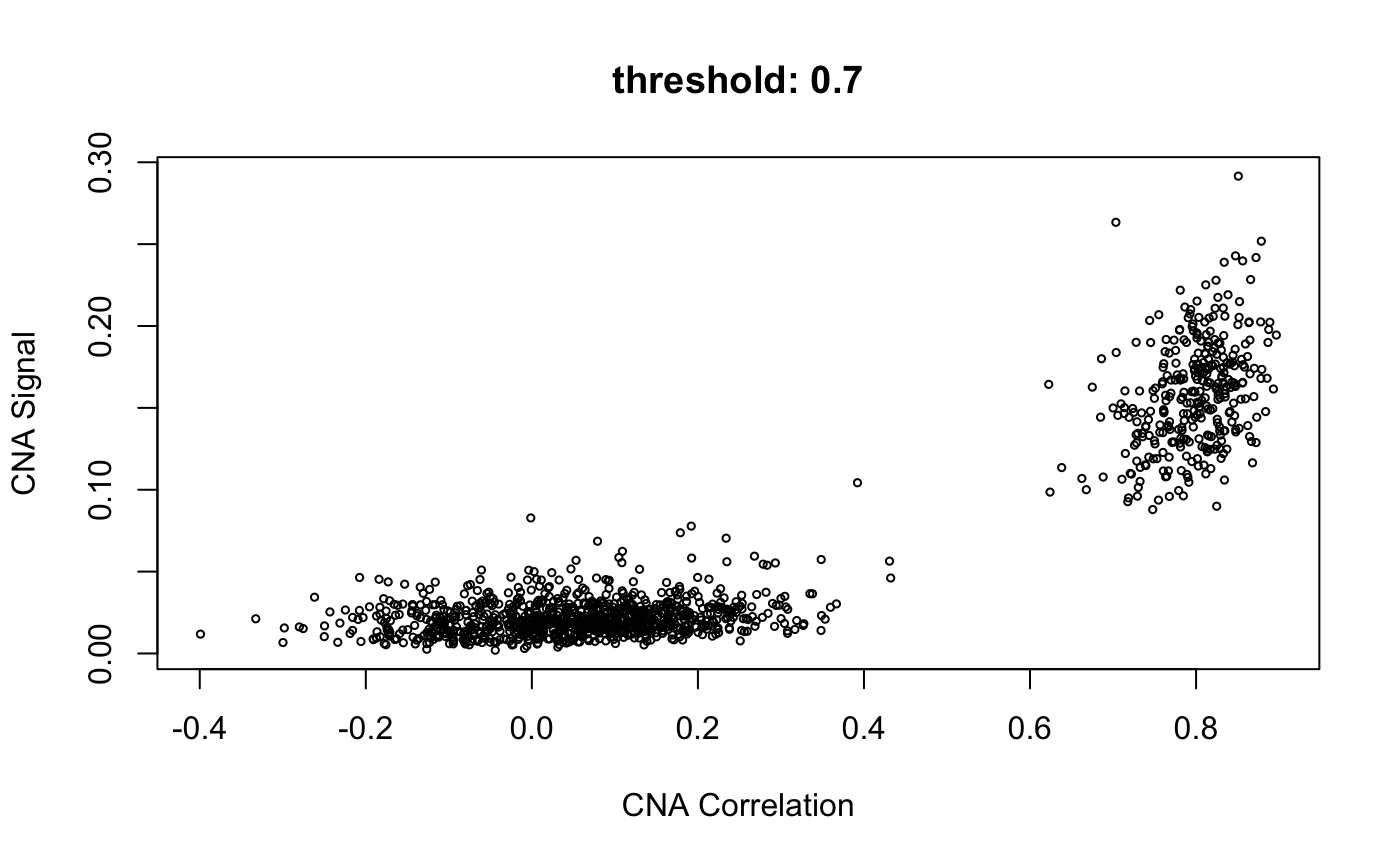
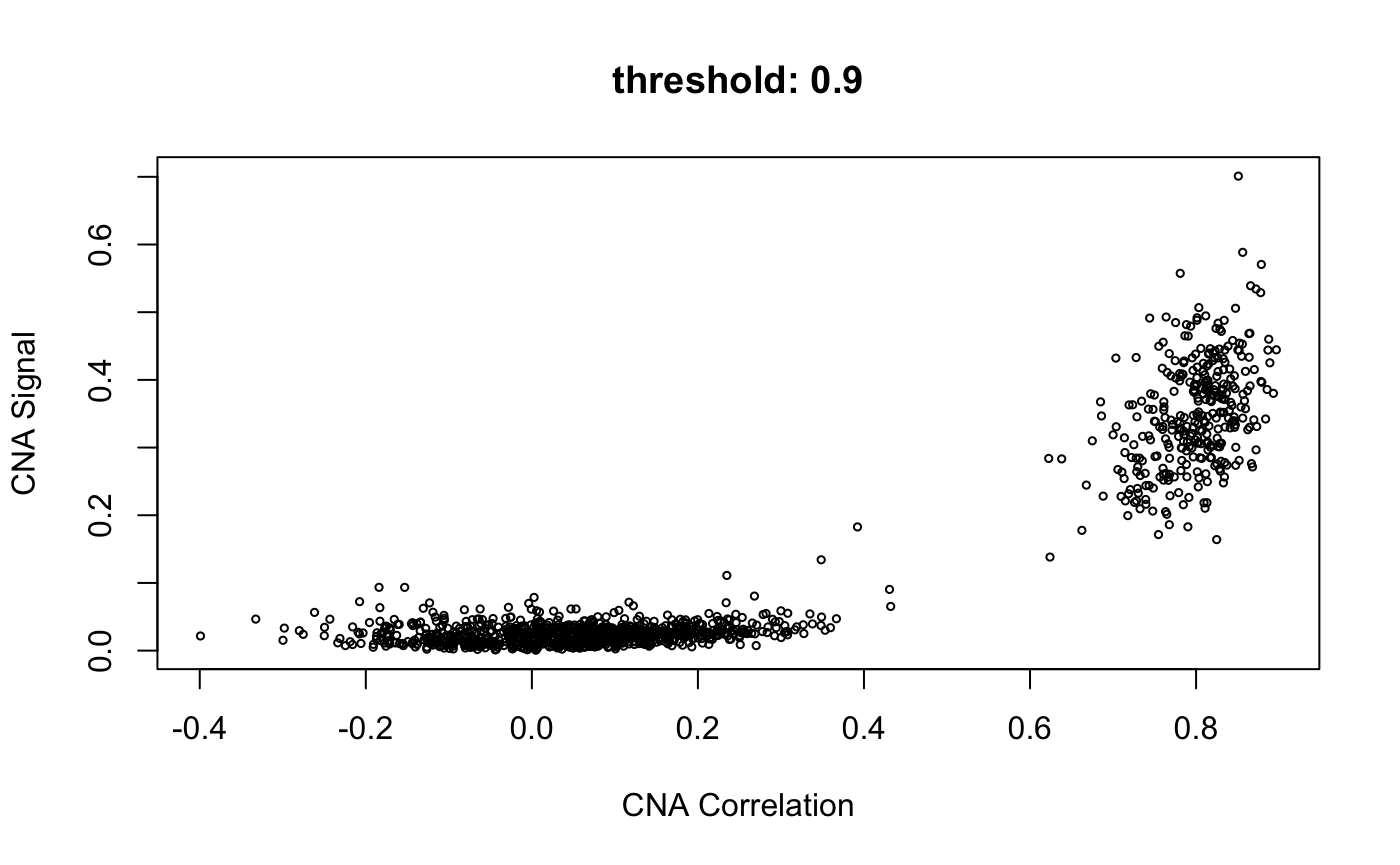
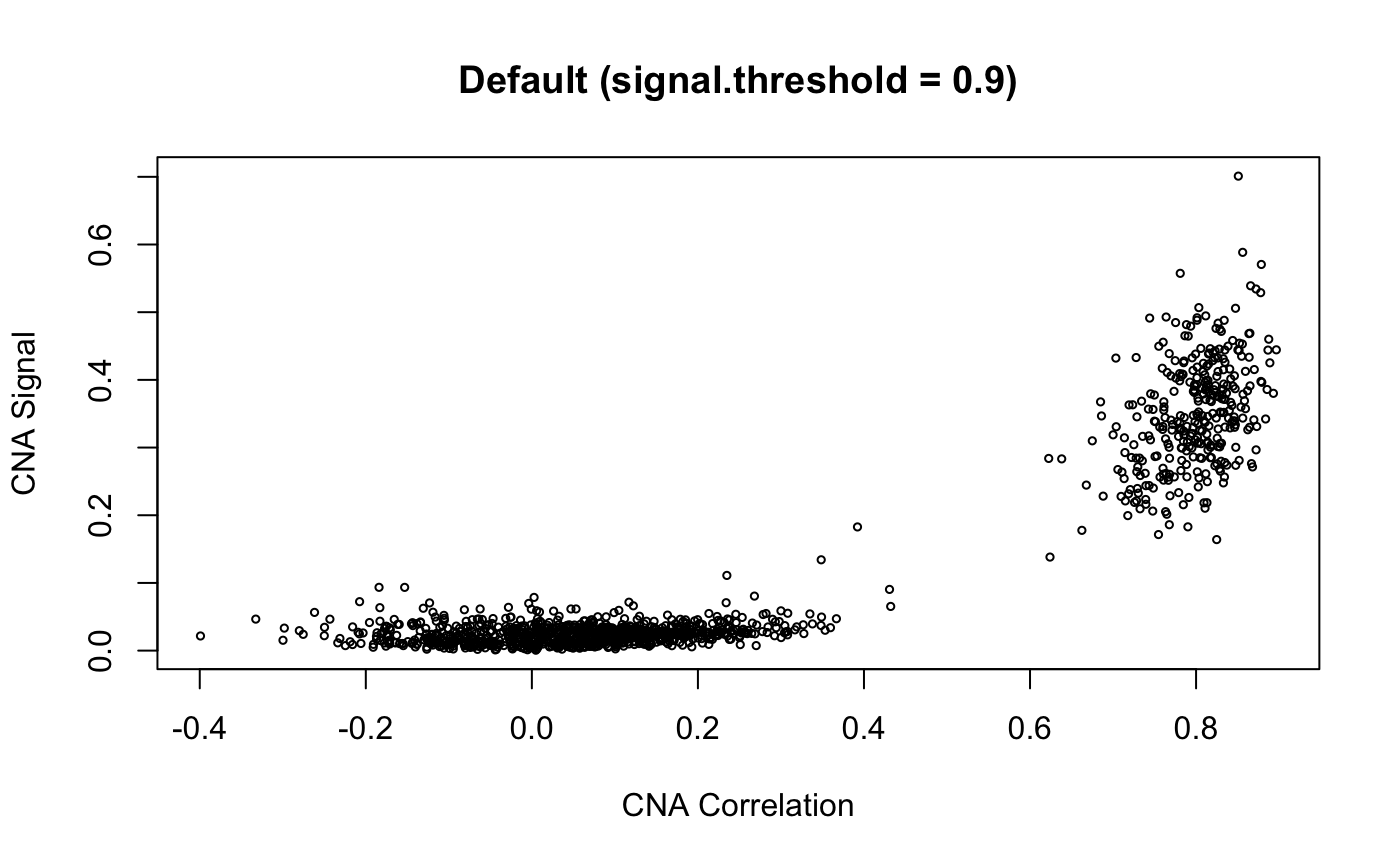
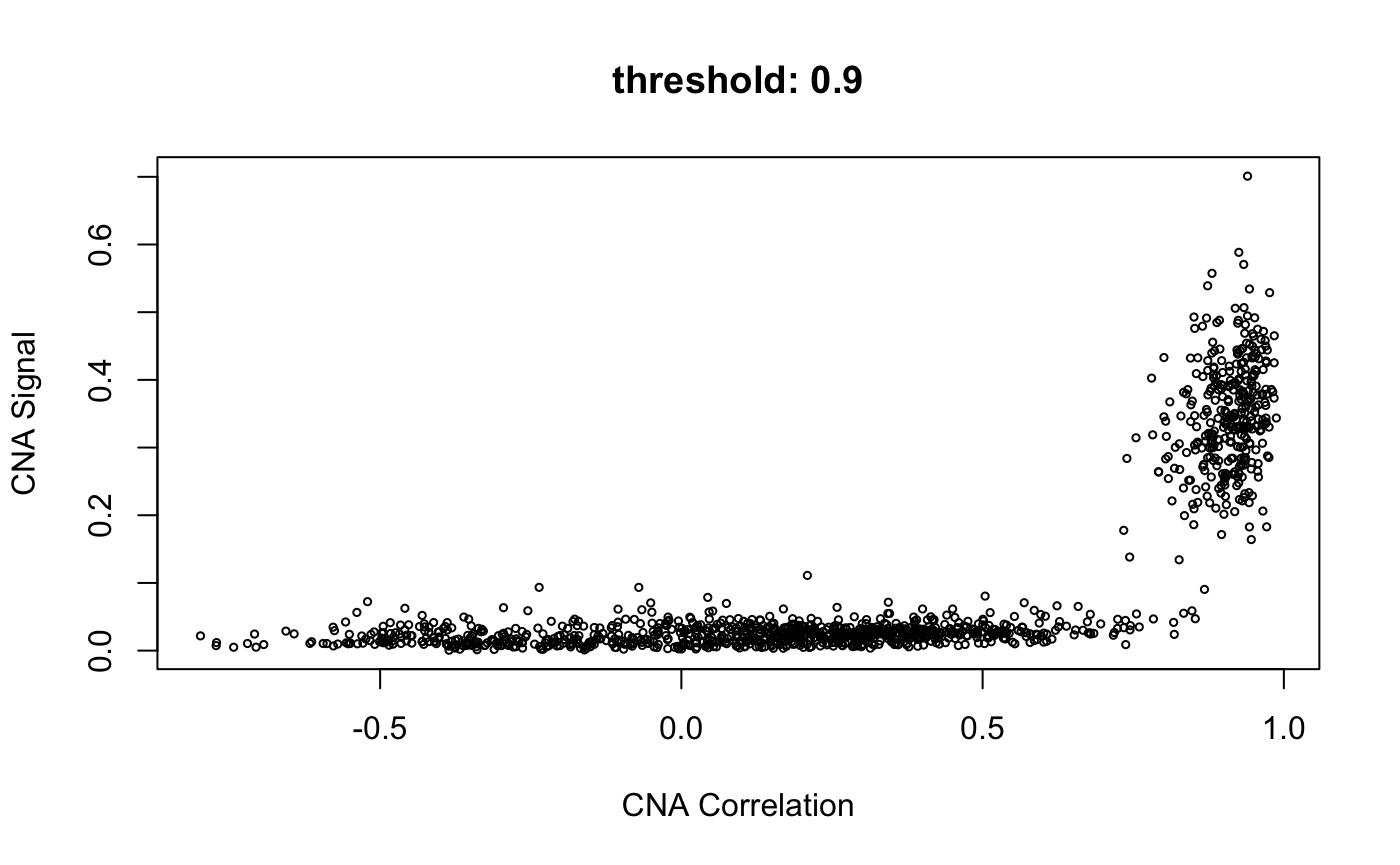
cnaScatterPlot(cna = cna, threshold = 0.9, samples = 'MGH125', excludeFromAvg = unlist(refCells), main = "threshold: 0.9, samples:'MGH125', excludeFromAvg")
cnaScatterPlot(cna = cna, threshold = 0.9, samples = 'MGH125', excludeFromAvg = unlist(refCells), main = "threshold: 0.9, samples:'MGH125', excludeFromAvg", group = unlist(refCells))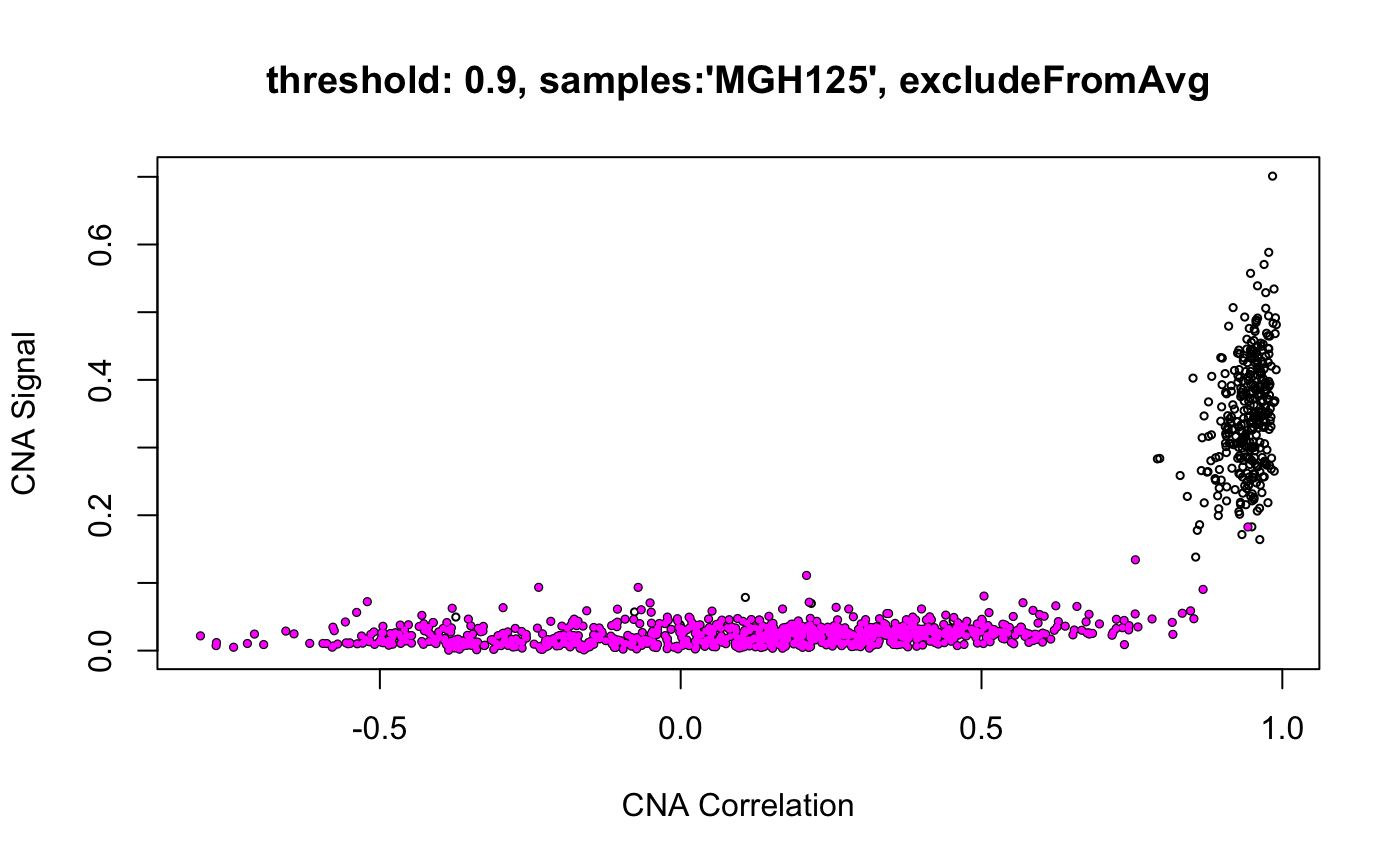
threshold finds top genes with cnaHotspotGenes()
When the <threshold> argument is set (to a value between 0 and 1) as above, infercna::cnaHotspotGenes() is called, and does two things:
Compute CNA signal values for each gene
-
Return genes with values of CNA signal in the top nth quantile
- where n is the value in threshold
Find malignant cells with findMalignant()
infercna::findMalignant() provides a first attempt at identifying malignant (and non-malignant) cells in the data. It fits bimodal distributions to the cells’ cnaSignal() and cnaCor() values, respectively. If two modes are found for each parameter, these are cross-checked and – if compatible –, joined and returned. If any of these steps fail, the return value is FALSE.
args(findMalignant)
#> function (cna, threshold = NULL, cor.threshold = threshold, signal.threshold = threshold,
#> bySample = FALSE, samples = NULL, sep = "-|_", mode.overlap = 0.05,
#> excludeFromAvg = NULL, cor.bySampling = TRUE, signal.bySampling = TRUE,
#> nsamples = 10000, ...)
#> NULLModes = findMalignant(cna, signal.threshold = .9, samples = 'MGH125')
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Found after 121 tries
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Success!
#> Found after 166 tries
#> removing 8 cells that were found in both modesFind genetic subclones
args(findClones)
#> function (m, prob = 0.95, coverage = 0.8, mode.size = 10, clone.size = 3,
#> by = "chr", bySampling = FALSE, nsamp = 2000, force.tries = FALSE,
#> verbose = FALSE, ...)
#> NULL
cnaCancer = cna[, Modes$malignant]
L = Map(filterGenes, value = c(2, '4p', 15), attribute = c('chr', 'arm', 'chr'), MoreArgs = list(cnaCancer))out = sapply(L, fitBimodal, assign = T)
#> Success!
#> Success!
#> Less than 80% of obs. could be assigned to a mode.out[[3]] = fitBimodal(L[[3]], assign = T, bySampling = T, nsamp = 300)
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Success!
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Success!
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> Success!lengths(out)
#> 2 4p 15
#> 2 2 2
sapply(out, lengths)
#> 2 4p 15
#> a 58 25 51
#> b 234 279 241
expandToClones(out, greaterThan = 3) %>% lengths %>% as.matrix
#> [,1]
#> 15.b 6
#> 2.a--15.a 6
#> 2.a--15.b 7
#> 2.a--4p.a--15.a 6
#> 2.a--4p.a--15.b 10
#> 2.a--4p.b--15.a 8
#> 2.a--4p.b--15.b 14
#> 2.b--4p.b 27
#> 2.b--4p.b--15.a 24
#> 2.b--4p.b--15.b 175
#> 4p.b--15.a 5
#> 4p.b--15.b 20findClones(cna[, Modes$malignant], by = 'chr') %>% lengths
#> At least one mode contains < 10 obs.
#> Success!
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> Less than 80% of obs. could be assigned to a mode.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.
#> At least one mode contains < 10 obs.Additional featuers
Split, Filter and Order vectors or matrices by Chromosome (Arms)
genesByChr = splitGenes(rownames(m), by = 'chr')
lengths(genesByChr)
#> 1 2 3 4 5 6 7 8 9 10 11 12 13 14 15 16 17 18
#> 844 593 502 295 432 391 473 297 313 262 458 466 153 269 240 354 490 115
#> 19 20 21 22 X Y
#> 549 238 86 186 305 5
datByChr = splitGenes(m, by = 'arm')
names(datByChr)
#> [1] "1p" "1q" "2p" "2q" "3p" "3q" "4p" "4q" "5p" "5q" "6p"
#> [12] "6q" "7p" "7q" "8p" "8q" "9p" "9q" "10p" "10q" "11p" "11q"
#> [23] "12p" "12q" "13q" "14q" "15q" "16p" "16q" "17p" "17q" "18p" "18q"
#> [34] "19p" "19q" "20p" "20q" "21p" "21q" "22q" "Xp" "Xq" "Yp" "Yq"