1 Load data
#> 6 x 6 sparse Matrix of class "dgCMatrix"
#> MGH136-P1-A02 MGH136-P1-A03 MGH136-P1-A05 MGH136-P1-A08 MGH136-P1-A09
#> A2M . . 1.578214 . .
#> AAAS 2.347666 4.457988 . 3.9442962 2.684145
#> AACS . . . . .
#> AAGAB . . . 0.7215914 .
#> AAMDC 5.031924 . . . .
#> AAMP . 5.272173 4.167759 4.0865290 5.165992
#> MGH136-P1-A10
#> A2M .
#> AAAS .
#> AACS .
#> AAGAB .
#> AAMDC .
#> AAMP 5.8523482 Center expression matrix
"""
MGH136.cent = t(scale(t(MGH136),center=T, scale=F))
# Or with sweep, apply, etc.
"""3 Generate an ordered correlation matrix
cr.matrix = scalop::hca_cor(MGH136.cent)"""
cr.matrix = scalop::hca_cor(MGH136.cent)
dist.matrix = as.dist(1-cr.matrix)
tree = hclust(dist.matrix)
cell.order = tree$order
cr.matrix = cr.matrix[cell.order, cell.order]
"""4 Retrieve cell clusters with a specified k
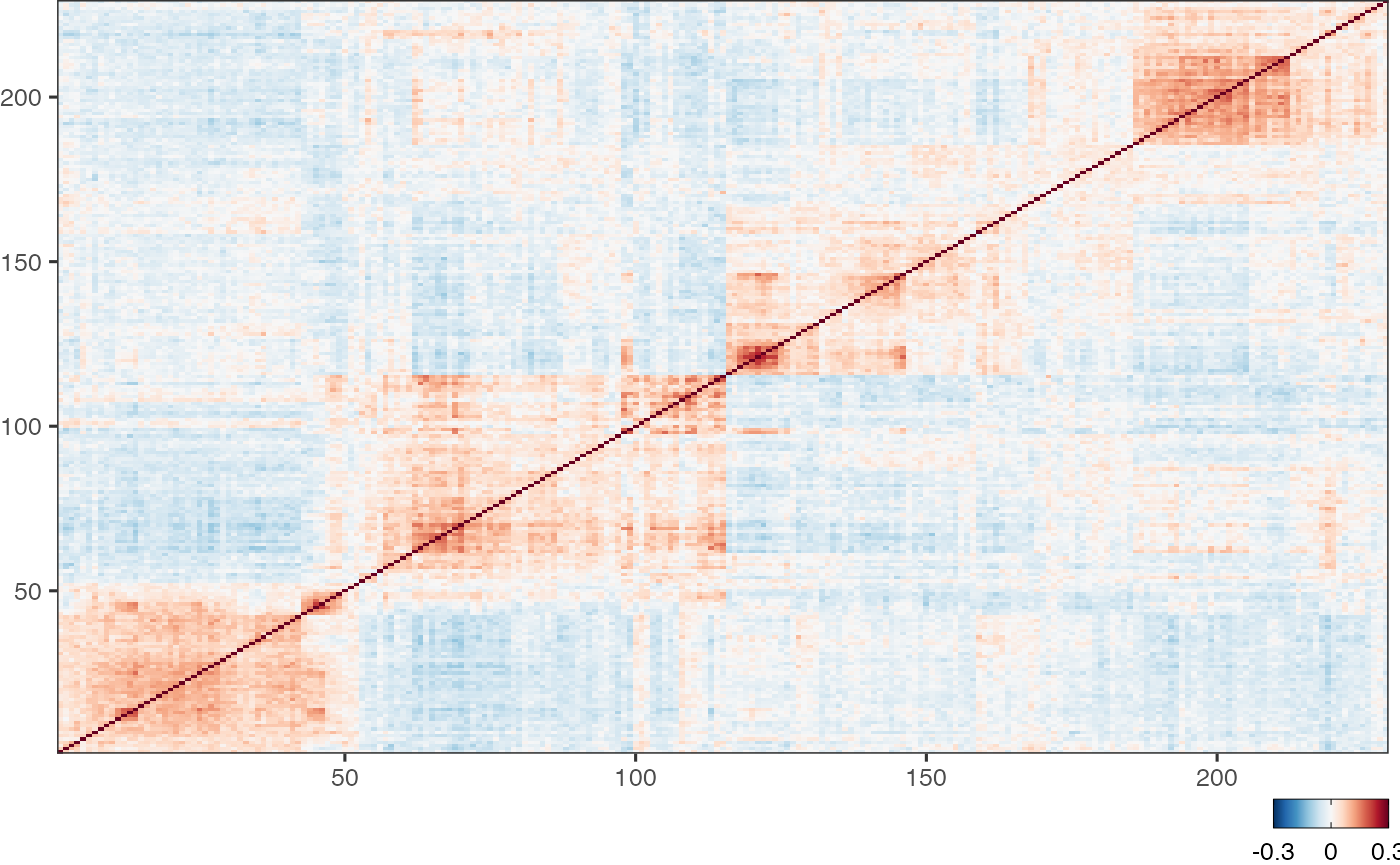
clusters_4 = scalop::hca_groups(
cr.matrix,
k=4,
cor.method='none',
min.size=0,
max.size=1
)"""
clusters_4 = stats::cutree(tree = tree, k = 4)
"""<img
programs_4 = scalop::dea(
MGH136.cent,
group=clusters_4,
center.rows=FALSE,
return.val='gene'
)
# ==========================
# Arguments (default values)
# ==========================
# lfc = log2(2) (or NULL)
# p = 0.01 (or NULL)
# pmethod = “BH”
# arrange.by = ‘lfc’ (or ‘p’)
# return.val = ‘lfc’ (or ‘p’ or ‘gene’)
lengths(programs_4)
#> 1 2 3 4
#> 223 361 149 182Let’s have a look at the top differentially expressed genes (DEGs) in each cluster:
scalop::ntop(programs_4, 100) %>%
as.data.frame %>%
scalop::setColNames(., c('NPC','CC','OPC','AC/MES'))#> NPC CC OPC AC/MES
#> 1 RND3 HMGB2 SPARCL1 GPR17
#> 2 STMN2 MAD2L1 VIM SIRT2
#> 3 CNR1 TYMS MT3 APOD
#> 4 ELAVL4 RRM2 CD9 BCAN
#> 5 EZR CDK1 ALDOC TMEM206
#> 6 DYNC1I1 ZWINT HEPN1 NEU4
#> 7 HMP19 UHRF1 PMP2 TNS3
#> 8 GNG3 KIAA0101 NDRG2 FIBIN
#> 9 TMEM161B-AS1 UBE2T HEY1 SERINC5
#> 10 KLHDC8A MLF1IP MT2A PLP1
#> 11 MGP PCNA GATM PHYHIPL
#> 12 ROBO2 UBE2C ATP2B4 CA10
#> 13 MIAT NUSAP1 FABP7 SCRG1
#> 14 OCIAD2 TOP2A WLS OLIG1
#> 15 LBH GMNN SLC1A3 SLC1A1
#> 16 ACTL6B PTTG1 NTRK2 HRASLS
#> 17 SYT4 MCM2 ZFP36L1 EPHB1
#> 18 FNBP1L CENPK GFAP TUBB4A
#> 19 SCG2 BIRC5 MLC1 TNR
#> 20 ENO2 CKS2 TTYH1 SOX8
#> 21 NREP MCM7 ZFP36 SMOC1
#> 22 TUBB2A RRM1 CLU AMOTL2
#> 23 BSCL2 MCM5 BBS2 LMF1
#> 24 CD24 CKS1B ATP1A2 RAB33A
#> 25 SEZ6 FAM64A CST3 FAM5B
#> 26 TAGLN3 TPX2 B2M TMEFF2
#> 27 THSD7A CCNB2 C1orf61 EPB41L2
#> 28 KLC1 HIST1H4C PON2 CNP
#> 29 PLK2 SMC4 ITM2C RTKN
#> 30 NSG1 KIF22 ATP1B2 CCSER2
#> 31 DLX5 RFC3 IL1RAP THY1
#> 32 MAP1B C19orf48 RHOC LIMA1
#> 33 NPPA ECT2 PPAP2B WASF1
#> 34 SEZ6L RNASEH2A PLAT AFAP1L2
#> 35 DCX GINS2 MASP1 MSMO1
#> 36 KIF5C ORC6 SAT1 P2RX7
#> 37 SNAP25 DNAJC9 PLOD2 LPPR1
#> 38 RBFOX2 PKMYT1 SLITRK2 PDE4B
#> 39 PKIA TMPO FOS ANGPTL2
#> 40 STMN4 CCNA2 SERPINE2 FYN
#> 41 SOX11 MELK IFI6 PSAT1
#> 42 CALM1 KPNA2 LGALS3 SOX6
#> 43 BLCAP GGH ATP13A4 SGTA
#> 44 TCF4 CCNB1 FKBP9 UGDH
#> 45 ROBO1 NUF2 APOD UGT8
#> 46 BTBD17 MCM3 TRIM9 TMEM123
#> 47 CDK6 H2AFZ CNN3 FAM110B
#> 48 CAMK2N1 FANCI SPARC TNK2
#> 49 DDAH2 CDCA5 LAMA4 ZDHHC9
#> 50 TUBB3 PBK P4HA1 SEMA5B
#> 51 PAK3 RFC4 ANXA2 TSPO
#> 52 RPH3A TACC3 PCDH9 VCAN
#> 53 DPYSL5 DUT EPAS1 IDI1
#> 54 CD200 CDKN3 BCHE SGK1
#> 55 SERINC1 NCAPG2 CALD1 GTF3C6
#> 56 CPE HMGN2 DTNA TCF12
#> 57 NDRG4 PRC1 HLA-C C16orf80
#> 58 NAPB CENPM LGALS3BP SEMA5A
#> 59 SRGAP3 BUB1B TIMP1 ANP32B
#> 60 ACOT7 DEK TIMP4 NKAIN4
#> 61 SPOCK1 CENPW SUMF2 TSC22D4
#> 62 PTPRO TUBA1B IGFBP5 OMG
#> 63 SSBP2 SMC2 MIR100HG ATCAY
#> 64 LMO1 TEX30 NRN1 PGRMC1
#> 65 MLLT11 ANP32E RNF180 GPD1
#> 66 APP VRK1 PTN LHFPL3
#> 67 PROX1 CHEK1 PROM1 KANK1
#> 68 SATB1 CENPF TAGLN2 MPZL1
#> 69 NFIA NDC80 EDNRB SNX22
#> 70 MMADHC PHGDH HLA-A SLC44A1
#> 71 SLC38A1 NUDT1 ANXA5 SCP2
#> 72 MIR100HG RFC2 SMOX PFN2
#> 73 SCG5 TMSB15A TAOK3 ZEB2
#> 74 LOC150568 ASF1B GPR56 CRB1
#> 75 GSTA4 DHFR JUNB DGCR2
#> 76 ARL4C FEN1 PTPRZ1 FXYD6
#> 77 ATP1A3 CENPN LAMB2 NME1
#> 78 PAM SNHG1 HLA-B ALG3
#> 79 ID4 FBXO5 PEA15 TNFRSF21
#> 80 LOC100506421 SPAG5 FOSB PAPSS1
#> 81 TRIB2 LIG1 MSI2 XPNPEP1
#> 82 ATAT1 EZH2 ADM RIN2
#> 83 STXBP1 CENPH TNIK MYC
#> 84 SOX4 CCNE2 RASSF4 TMEM258
#> 85 ENC1 DTL S100A16 SRPRB
#> 86 DRAXIN RACGAP1 HTRA1 MAP1A
#> 87 STMN1 RAD51AP1 CSPG5 CADM2
#> 88 ARL4D SNRPB HSPA2 TAGLN3
#> 89 TULP4 SLBP LITAF DLL3
#> 90 BASP1 CKAP2 SLC2A3 LRRN1
#> 91 CRABP1 TUBB4B SPATA6 INSIG1
#> 92 SNRPN TK1 SOX9 CRIPT
#> 93 DYNLT1 MZT1 SLC35F1 SHD
#> 94 HN1 KNSTRN TXNIP GPX1
#> 95 ASIC4 MCM4 CRYAB ERBB3
#> 96 ATP1B1 CSRP2 VAMP3 REC8
#> 97 UCHL1 CDCA7 N4BP2L2 TMCC1
#> 98 SNURF CDCA7L DDR1 PLXNB3
#> 99 ATP6V0A1 GGCT TMEM255A NDUFB2
#> 100 LINC00461 CDC20 S100A6 ALCAM6 Retrieve cell clusters with no cutoff
Let’s revisit step 4. Can you see the problems with this method?
MGH136clearly showed four clusters of cells, but many tumours would not be so structured: the correlation maps would look be messy, with no obvious number of clusters to choose.This method would limit us to working with mutually exclusive clusters of cells, whereas in reality we would prefer some degree of redundancy, since biology is redundant, after all!
To address these points, we can instead retrieve clusters at many values of k (a.k.a. heights on the dendrogram) and thereby avoid selecting clusters at arbitrarily defined cutoffs.
# The k argument now takes a value of NULL
clusters_all = scalop::hca_groups(
cr.matrix,
k=NULL,
cor.method ="none")
### ADD EQUIVALENT CODE IN BASE R